New technique shows centromeres are hotspots for DNA breaks
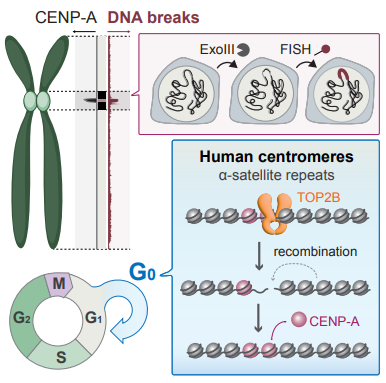 Centromeres are critical regions of the eukaryotic genome that provide structural platforms for assembling kinetochores, which mediate chromosome segregation in cell division. Centromeres consist of tandemly repeated DNA sequences, known as satellite repeats, and are defined functionally through the presence of CENP-A histone variant for kinetochore attachment. For decades, centromeres have been known to evolve very quickly, with satellite repeats differing drastically between closely related species. This is in contrast to the highly conserved role of centromeres across eukaryotes and has been a mystery.
Centromeres are critical regions of the eukaryotic genome that provide structural platforms for assembling kinetochores, which mediate chromosome segregation in cell division. Centromeres consist of tandemly repeated DNA sequences, known as satellite repeats, and are defined functionally through the presence of CENP-A histone variant for kinetochore attachment. For decades, centromeres have been known to evolve very quickly, with satellite repeats differing drastically between closely related species. This is in contrast to the highly conserved role of centromeres across eukaryotes and has been a mystery.
Here, Saayman et al. shed light on this long-standing question. They used publicly available short-read sequencing databases to map the occurrence of DNA breaks onto the recently released complete human genome, gaining the first insight into centromeres as hotspots of DNA breaks. They complemented this approach with a new microscope-based technique they developed, exoFISH. Combining these techniques, the team demonstrated that DNA breaks occur on centromeres both in dividing and non-dividing cells. Focusing on centromeres in non-dividing cells, they showed that the breaks are likely due to Topoisomerase IIβ, an enzyme that controls the topologic states of DNA, and that RAD51, a key player in Homologous Recombination, helps resolve these breaks. This observation provides evidence for the theory that homologous recombination plays a crucial role in centromere rearrangements and maintenance, likely through driving CENP-A localization to the centromere.
One of the most striking findings from the paper is the observation of high rates of centromeric DNA breaks in non-dividing, quiescent cells. As Prof. Esashi explained herself, “This study opens so many questions in this understudied area of biology, relating many of those cells in resting conditions in our body, such as lymphocytes, oocytes, and dormant cancer cells.” “I am excited to learn more about what is happening at centromeres during quiescence and their impact on genome evolution and human disease in the future.”
- Written by Lena Duma (Ivan Ahel lab)
Explore more
Read the paper
Centromeres as universal hotspots of DNA breakage, driving RAD51-mediated recombination during quiescence.
Esashi Group
The Esashi group investigates the mechanisms that drive changes in the genome during cell growth and dormancy.
Cell and Developmental Biology
Several Dunn School groups investigate the mechanisms underlying a range of important developmental and cellular processes such as signalling, transcriptional control, cell division, protein trafficking, and genome maintenance.