“The Unknome”: The database of neglected proteins created by Dunn School researchers
Humanity had to wait about 150 years from the discovery of genes for the whole human genome to be sequenced at the start of the 21st century. However, with the world of information about the genome now at our fingertips, it has become increasingly apparent how much there is still left to uncover. Once undergraduate lab partners, Matthew Freeman and Sean Munro have teamed up again to show that the quest to fully understand our genome is far from over.
In their project, the authors have developed a new bioinformatic method to quantify how much is known about each gene. They then combined this ‘known-ness’ score with existing sequence information to rank all genes by how much (or little) is known about them. This was developed into the ‘Unknome Database’, which highlights the most widely conserved genes about which essentially nothing is known. Additionally, a user can customise their database by modifying a protein’s knowness score based on their own set of criteria.
The validity of the system could be seen when the top 10 genes with the highest knowness scores were confirmed as having well-established roles in cell function and development. In contrast, 1,723 human genes out of 19,664 achieved a knowness score of 1 or less – implying that nothing is known about them. To demonstrate the usefulness of their database, the authors picked a set of 358 such genes, with orthologs in both humans and fruit flies, and used RNA interference to knock them down in Drosophila. A quarter of them were found to be lethal after deletion, and the removal of a further quarter showed marked changes in the fly’s phenotype. Partial or tissue-specific knockdowns helped to confirm that many of these genes participate in a wide range of biological processes – from development, through locomotion and fertility, to resilience to stress. These results confirmed that the genes of unknown role do indeed have essential functions and, in the words of Matthew Freeman, ‘do not deserve their neglect’. Although the proof of concept was done in flies, the overall goal of the project is to identify the human genes relevant to new biology and medical opportunities.
Despite the explosion of scientific data, the authors find that the “Unknome” is shrinking only slowly. The analysis of publication trends reveals that funding bodies tend to preferentially support projects concerning proteins with proven clinical importance, as well as projects with lower perceived risk. Further, scientific factors have also been proposed, such as lack of specific antibodies, or uncertainty of protein levels in the cell, exacerbating what the authors elegantly call the “neglect of unknown”. As Matthew Freeman explains: ‘everything points to these genes having important biomedical function. It is crucial that scientists, funders and companies are bold and imaginative. There is a huge incentive for us to start working on the really unknown parts of the human proteome, and we hope that this publicly available resource will help people choose exciting projects that are genuinely novel.’
Written by Aleksandra Pluta (Murphy lab) @aj_pluta
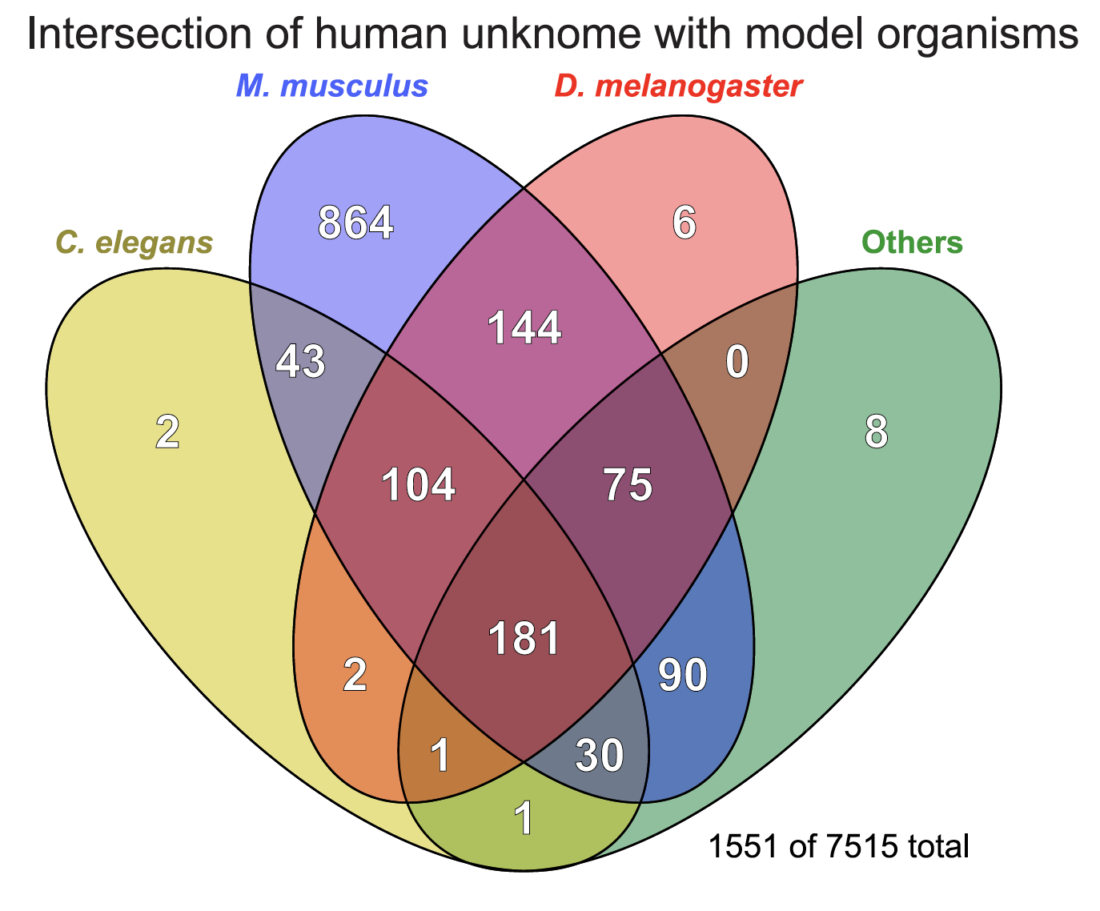
A Venn diagram showing the distribution of genes across different species, from clusters of knownness score <2.0 and which contain at least 1 human protein.
Read the paper
Functional unknomics: Systematic screening of conserved genes of unknown function.
Freeman group
Investigating the interface between membrane proteins, the cell biology of signalling, and mechanisms of human disease
Cell and Developmental Biology
Several Dunn School groups investigate the mechanisms underlying a range of important developmental and cellular processes such as signalling, transcriptional control, cell division, protein trafficking, and genome maintenance.