How Breast Cancer Hijacks a Natural Enzyme to Boost Mutations
This collaboration from the Gromak (University of Oxford), Harris (University of Texas Health Science Center at San Antonio), and Miller (University of Texas at Austin) groups, demonstrates for the first time that A3B interacts with R-loops. R-loops are three-stranded structures composed of an RNA-DNA hybrid and displaced ssDNA. These structures form naturally during transcription but their misregulation is linked to various diseases, including cancer.
Here, the authors demonstrate a previously unrecognised role for A3B in regulating R-loops. They further show that A3B binding to R-loops promotes mutations at these sites, through a genome-wide mapping of A3B-associated R-loops and A3B signature mutations in breast cancer. Hence, they unveil a novel mechanism for R-loop-associated mutagenesis in cancer.
Importantly, this research published in Nature Genetics highlights the potential for targeting R-loops in new cancer therapies. Drugs that drive R-loop accumulation, such as topoisomerase inhibitors, already approved by FDA, could lead to excessive mutation in cells that overexpress A3B, allowing the selective targeting of cancer cells.
Written by Chiara Beghe and Helena Harpham
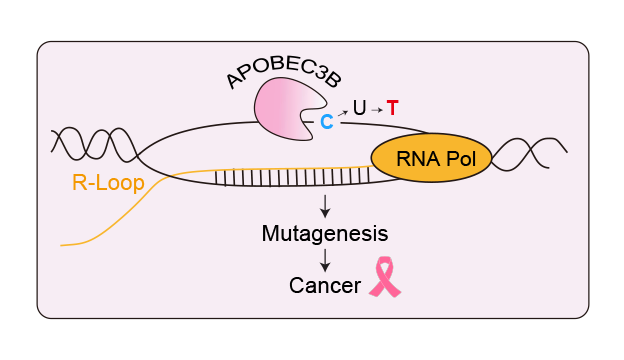
Image credit: Kyle Miller (University of Texas at Austin).
Explore more
Read the paper
Read about how APOBEC3B proteomics reveal interactions with a surprising number of R-loop factors.
Gromak Group
Studying the role of non-canonical RNA/DNA structures called R-loops in health and disease.
RNA and Gene Expression
Several Dunn School groups use a range of molecular biology approaches to investigate the fundamental mechanisms underlying transcription, transcriptional gene silencing, and RNA processing.