dCas9 roadblocks paving new ways for CRISPRi use
Our approach to genome editing changed drastically in 2012, with the discovery and implementation of the CRISPR-Cas9 system. CRISPR, short for Clustered Regularly Interspaced Short Palindromic Repeats, refers to unique sequences in the bacterial genome, which act as its immune system. Cas9 is an endonuclease protein that associates with CRISPR-derived RNAs, that in effect target formation of DNA breaks at very precise locations. Although primarily known as a tool for gene editing, it is becoming abundantly clear that the CRISPR-Cas9 mechanism can be used for a variety of other research applications – from the detection of specific nucleic acid sequences, to the control of gene expression.
The latter involves the use of enzymatically “dead” Cas9, dCas9, which continues to bind specific DNA sequences with the use of CRISPR guide RNAs (gRNAs), but it now lacks its nuclease activity. It can be linked to effector domains such as in CRISPRa and CRISPRi – systems for, respectively, activating or inhibiting transcription. However, despite the plethora of available gRNAs for targeting distinct points within the genome, the full picture of the effects of dCas9 binding remains somewhat blurry. Previous investigations have shown that dCas9 bound to the gene transcription start site blocks its expression, but little was known about the impact of targeting dCas9 across downstream positions on the gene.
In their new work, Inna Zukher et al. employed the dCas9-KRAB CRISPRi suppression system to corroborate that binding dCas9 to the 5’ end of a gene significantly reduces its expression. Correspondingly, targeting it downstream of the polyadenylation signal (PAS) causes a blockade for transcriptional elongation, as judged by a decrease in RNA readthrough. Natural anti-termination effects, for instance stress-induced, can still counteract early termination caused by CRISPRi. Pol II pausing is seen to occur just upstream of the DNA-bound dCas9, and is further associated with increased Thr4 phosphorylation in the polymerase’s C-terminal domain, which eventually triggers termination. Although the KRAB domain recruits histone methyltransferases and increases the local H3K9me3 levels, the Pol II stalling effect seen in CRISPRi-induced early termination was found not to be dependent on repressive chromatin marks, but rather on the dCas9 roadblock itself.
Importantly, the data obtained in the investigation has underlined that the CRISPRi roadblock is orientation specific. Although dCas9 can target both DNA strands equally well, it does not have any pronounced transcriptional effects when bound to the template strand outside of the transcription start site region. These results could be of critical use when designing dCas9 experiments where minimal transcriptional disturbance is required without compromising on binding efficacy. The exact consequences of dCas9 roadblocks on early termination were further analysed by targeting the protein to different sites around the PAS. It was shown that when paired with an antisense guide, dCas9 acts as a sturdy roadblock to the elongation complex, but only transiently stops Pol II progression. The resulting collision between the polymerase and dCas9 culminates in earlier termination of transcription, but with no change to overall gene expression if there is an active PAS upstream of the block. However, if the PAS is nowhere close by, the transcript fails to get polyadenylated and gets degraded, which in turn suppresses gene expression. The site of dCas9 binding is also important when the gene has two active PAS, in which case alternative polyadenylation events are employed. Finally, the transient nature of the block is further verified by its lack of effect on alternative splicing, which is usually influenced by Pol II kinetics.
Taken together, the findings collected by Zukher and colleagues shed a new light on the elusive nature of CRISPRi interactions with Pol II and the elongation complex, and so help us better understand how the dCas9 roadblock might affect gene expression based on the gene site it targets. Ultimately, gaining better knowledge of this elegant system will help achieve greater robustness in the ever-expanding catalogue of CRISPR experiments.
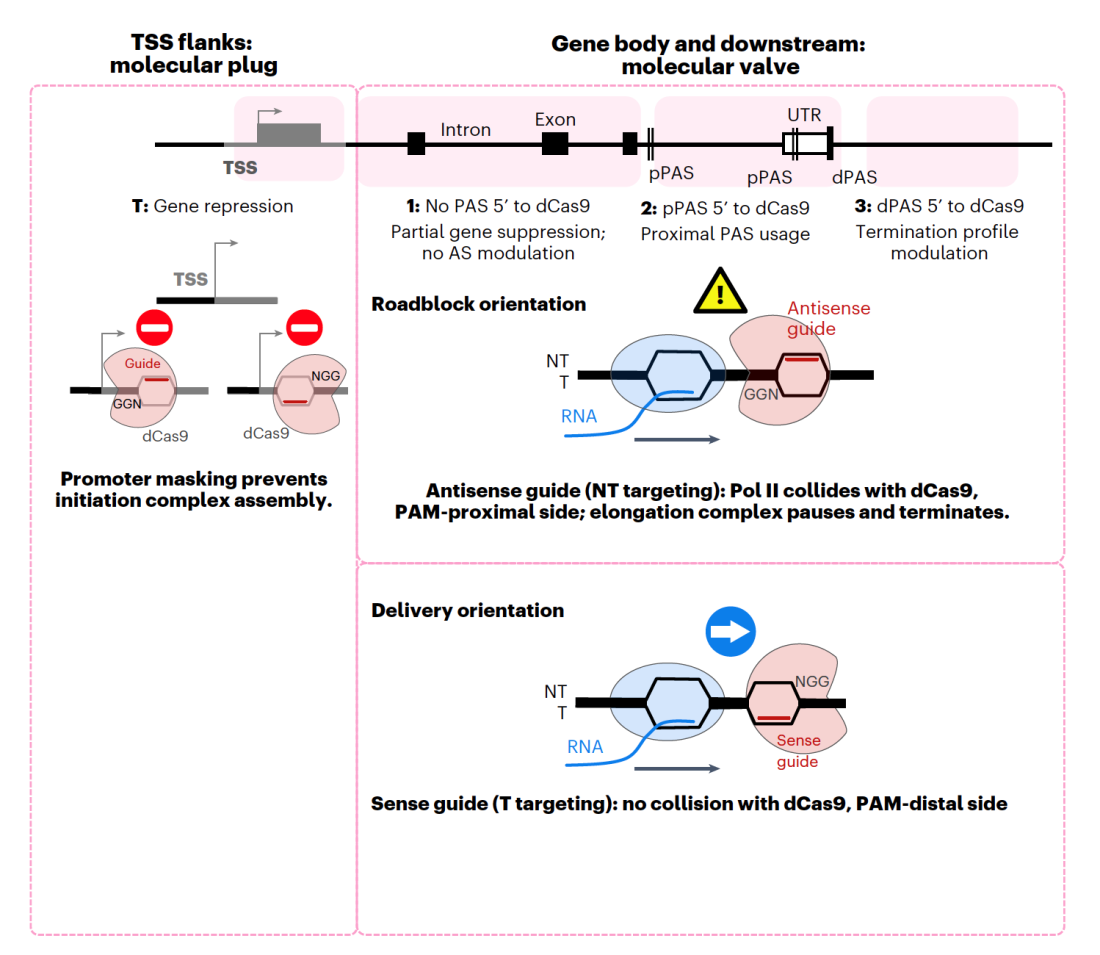
Differential effects of a dCas9 roadblock across a gene.
Written by Aleksandra Pluta (Murphy lab) @aj_pluta
Explore more
Read the Paper
Proudfoot Group
Studying the molecular mechanisms that define the extent of transcription units generated by RNA polymerase II across mammalian genomes.
RNA & Gene Expression
Several Dunn School groups use a range of molecular biology approaches to investigate the fundamental mechanisms underlying transcription, transcriptional gene silencing, and RNA processing.